Upgrading Florida Tech's CAPP Reports (Part 1)
Note: This two-part post was originally written as a single Jupyter Notebook (download here).
The Problem
Florida Tech's course catalog is pretty good. It's got nice hyperlinking, helpful popups, and a reliable search system. It even includes degree requirements for various majors. Let's see a screenshot:
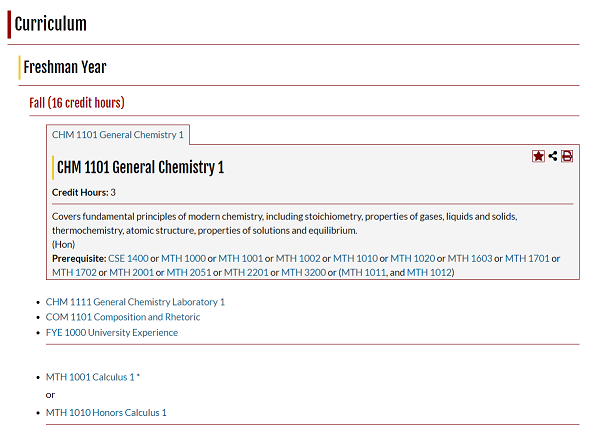
Cool! Not too hard to follow, and very nice and interactive.
But what if I want to check which of those degree requirements I'm meeting?
Florida Tech's PAWS system for students has a handy tool just for that -- CAPP Reports (Curriculum, Advising, and Program Planning). Students can generate "reports" that tell them which requirements they need to meet / have met so far. Let's see how it looks:
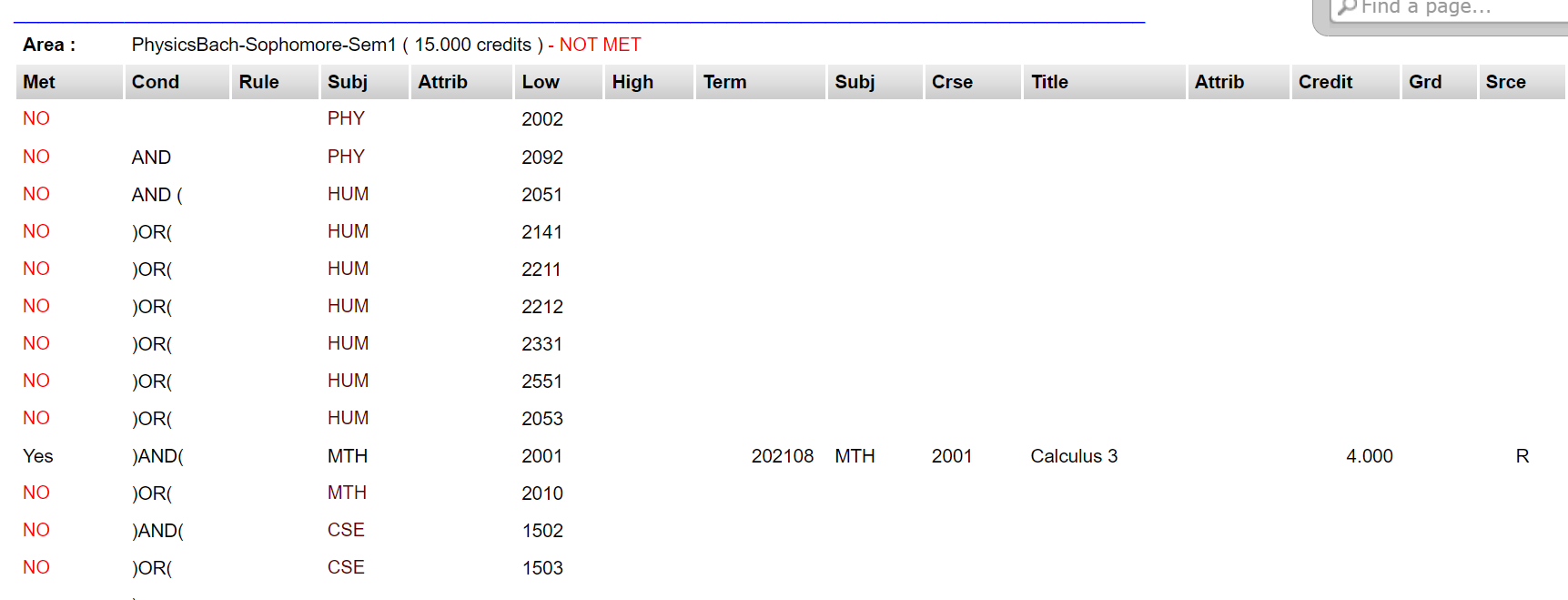
Bleh. And this is just what I could fit in a screenshot -- these tables go on for pages. They aren't linked to the course catalog, so while planning courses, you need to have multiple tabs open while switching back and forth, double-checking that you entered the CRN right.
What I'd like to have is something like this:
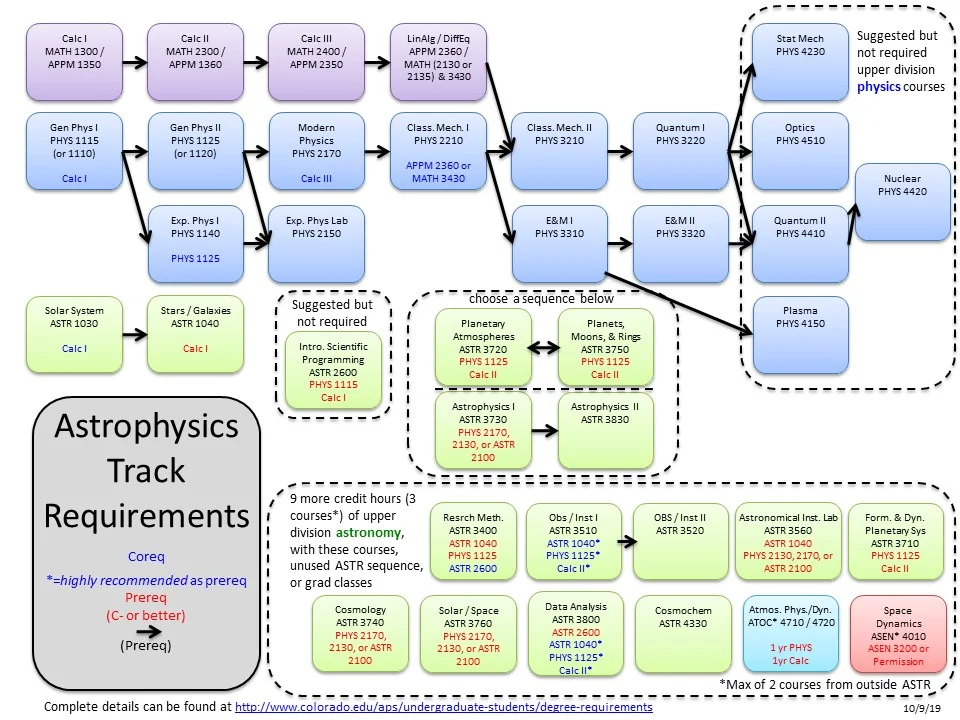 Courtesy of the University of Colorado
Courtesy of the University of ColoradoThe Solution
Yes, I could just draw a flowchart for the Physics major. This would be simple and easy. Unfortunately, I am a computer programmer, and consequently duty-bound to automate any menial task I encounter.
In this document I'll develop a procedure for transforming a CAPP report into a personalized, readable course dependency flowchart constructed from publicly available data.
First we'll need Florida Tech's course catalog.
Extracting the Catalog
The course catalog does not have an API, and is stored in its entirety on a series of webpages found at https://catalog.fit.edu/content.php?catoid=12&navoid=551. Download the .html files of all 26 of these pages and save them.
Combine the pages together. Open a linux shell (I'm on Windows, so I used WLS Ubuntu) and run:
ls -tQ *.html | xargs cat > concat.htmlThis page contains the course names, but not their descriptions, which are only shown when a course name is clicked on. Thankfully, this page does contain links to individual course pages with these descriptions on them. This command extracts the links to these locations (first grep gets lines, second gets links, then sed removes those pesky "amp;"s):
grep -e $'<td class=\"width\">. \t\t\t\t<a href=\"' concat.html | grep -Eo 'https://[^\"]+' | sed 's/\&/\&/g' > links.txtWe now have a list of links to every course description in the catalog (careful, these description pages can change frequently). Move links.txt to new, empty folder.
Back in linux, run:
(sorry Florida Tech servers)wget -i links.txtProcess the HTML files...
Extracting the Course Network (with Python)
Libraries
I'm managing my environment with Anaconda running Python 3.8. It might be annoying on windows. The Anaconda Navigator can help. distinctipy and pyvis aren't part of Conda, so you'll have to install them manually. Don't get pyvis confused with pyviz, which is an entirely different library (this cost me far too much time).
from os import walk
import matplotlib.pyplot as plt
import networkx as nx
import numpy as np
import pandas as pd
import re
from graphviz import Digraph
import holoviews as hv
from holoviews import opts
from pyvis.network import Network
from distinctipy import distinctipy
import math
import jsonThe Course Class (Class Class?)
Contains all data regarding a course, sans any fluff text after the course description but not in the course "requirements" section.
class Course:
def __init__(self):
self.page_num = 0
self.course_num = 0 # 1150
self.course_code = '' # 'EGN'
self.course_id = '' # 'EGN1150'
self.course_name = '' # 'Combined Machine Shop Certification'
self.credit_hours = '' # can be a range, like 1-3
self.description = ''
self.requirement = ''
self.prerequisites = []
self.corequisites = []
self.recommended = []
self.complements_courses = []
# the following three values aren't used till much later in this notebook
self.met = False # has this course been taken?
self.in_major = False # is this course listed in the major reqs?
self.req_attrs = []
# Possible values: CC HU SS LA Hon Q CL
self.tags = []
def print_blurb(self):
print(self.course_code + ' - ' +
str(self.course_num) + ' ' +
self.course_name +
'\nCredit Hours: ' +
str(self.credit_hours))
print('Requirement: ' + self.requirement)
print('Corequisites: ' + ' '.join(self.corequisites))
print('Prerequisites: ' + ' '.join(self.prerequisites))
print('Recommended: ' + ' '.join(self.recommended))
print('This course complements: ' + ' '.join(self.complements_courses))
print('\n')Regex Helper Functions
Parsing HTML with regex is going to be messy. It's usually a BIG no-no, but I'm allowed do it here because I know the exact format of all the HTML I'm working with. The following code is very specific to the data I have. It won't be pretty, but if I wanted pretty, I'd be using BeautifulSoup or some other HTML parser.
# searches target for a substring in between start and end
# this code actually isn't specific to this project at all... :P
def in_between(target, start, end):
return re.search(start + '(.*)' + end, target).group(1)# removes all .HTML tags (simple)
def remove_tags(target):
return re.sub('<[^<]+?>', '', target)def reformat_list(l):
# add spaces so that the parenthesis are recognized seperate from coursenames
# have fun reading this line lmao
s = re.sub('\(', '( ', re.sub('\)', ' )', ' '.join(l))).strip()
# change all 'BME 3081' to 'BME3081'
o = re.sub('(?<=[A-Z]{3})\s(?=[0-9]{4})', '', s).split(' ')
# remove empties
return list(filter(None, o))walk() is a very handy function.
def get_filenames(path):
f = []
for(dirpath, dirnames, filenames) in walk(path):
f.extend(filenames)
return fParsing HTML
This is where things start to get messy. This code only works on the 2021 course catalog I downloaded from Florida Tech, and will probably need to be updated if the catalog changes or is reformatted. I'm handling a lot of edge cases here. The only way definitive solution to this would be a ML-based approach (god forbid) or just access to the registrar's database itself.
Though if I had that, this process would look very different.
def parse_html(path_to_folder):
names = get_filenames(path_to_folder)
try:
names.remove('links.txt')
except ValueError:
pass
course_list = []
for html_path in names:
c = Course()
c.page_num = html_path[-5:]
f = open(path_to_folder + html_path, 'r', encoding='utf8')
lines = [line.rstrip() for line in f]
# We've already opened an HTML file, now we need to scan through it until we find the line with the course
# information on it.
# IMPORTANT: This doesn't always work. For MAR 6899, the data was broken by two newline characters for some
# reason. I manually corrected this in the file. Proper HTML parsing would fix this.
line = ''
for s in lines:
if '<p><h1 id=\'course_preview_title\'>' in s:
line = s[51:] # crop out initial junk
break
# CN and dept. code are always the title of the box
c.course_num = int(line[4:8]) # 4302
c.course_code = line[:3] # 'AVT'
c.course_id = c.course_code + str(line[4:8])
# split things up by linebreaks for manageability
sections = line.split('<br>')
c.course_name = sections[0].split('</h1>')[0][9:] # 'Aviation Safety Analysis'
# I'm removing the <p> tags because a couple of courses, namely PSY 6550 and
# MAR 6899, are formatted differently and have <p> tags in their HTML. What's up with those?
c.credit_hours = remove_tags(in_between(re.sub('<p>', '', sections[0]), '</h1><strong>', '<hr>'))[14:]
c.description = sections[0].split('<hr>')[1]
# Some of the descriptions say "complements [some other course]" or "builds on [course]"
# I use a catch-all here to grab those instances
if 'complements' in c.description.lower():
c.complements_courses = re.findall('[A-Z][A-Z][A-z] [0-9][0-9][0-9][0-9]', c.description)
#print(sections[1])
for i in range(1, len(sections)):
# remove tags, strip, and remove non-breaking space character escapes
sec_notag = re.sub(' ', '', remove_tags(sections[i]).lstrip())
# It's one of those lines that looks like "(CC) (HU/SS) (LA) (Hon)"
if re.sub('Hon', 'H', sec_notag).isupper():
tagcat = re.sub('\(|\)', ' ', sections[i])
# tagcat = re.sub('/', ' ', tagcat) # uncomment to remove the bipartite tags (like 'HU/SS')
# remove empty strings
c.tags = list(filter(None, tagcat.split(' ')))
elif sec_notag.startswith('Requirement'):
c.requirement = sec_notag.split(':')[1][1:]
elif sec_notag.startswith('Prerequisite'):
# get the junk out
sec_notag = re.sub(',', '', sec_notag)
sec_notag = re.sub('\xa0', ' ', sec_notag)
# ECE3331 has a typo in this section (no second space in 'ECE 3331or')
# fix those sorts of errors here
# before we sub out the "OR"s, we need to sub out the "cORequisites" (this created some errors, heh)
sec_notag = re.sub('Corequisite', ' CRQ ', sec_notag)
sec_notag = re.sub('or', ' or ', sec_notag)
sec_notag = re.sub('and', ' and ', sec_notag)
sec_notag = re.sub('/ {2,}/g', ' ', sec_notag)
# split the prerequisite requirements
# e.g. '(PHY 0000 and MTH 0001) or BIO 1111'.split(' ')
# remove all extraneous strings from array
prereq_cmds = sec_notag.split(':')[1].split(' ')[1:-1]
while not prereq_cmds[-1]:
del prereq_cmds[-1]
c.prerequisites = prereq_cmds
# corequisites are stored on the same line as prerequisites, parse them similarly
if 'CRQ' in sec_notag:
# slightly different slicing for formatting reasons
coreq_cmds = sec_notag.split(':')[2].split(' ')[1:]
while not coreq_cmds[-1]:
del coreq_cmds[-1]
c.corequisites = coreq_cmds
elif sec_notag.startswith('Recommended'):
sec_notag = re.sub(',', '', sec_notag)
sec_notag = re.sub('\xa0', ' ', sec_notag)
# this section has less consistent formatting, so we have to walk through it line-by-line
if ":" in sec_notag:
part = sec_notag.split(':')[1]
starr = re.findall('(?:to|or|and).[A-Z]{3}\s[0-9]{4}', part)
if len(starr) > 0:
if starr[0].startswith('to '):
starr[0] = starr[0][3:]
rec_cmds = []
for s in starr:
rec_cmds += s.split(' ')
c.recommended = rec_cmds
else:
# yeah, this is a possibility. For just one course. :|
c.recommended = [sec_notag]
c.complements_courses = reformat_list(c.complements_courses)
c.corequisites = reformat_list(c.corequisites)
c.prerequisites = reformat_list(c.prerequisites)
c.recommended = reformat_list(c.recommended)
course_list.append(c)
return course_listNetwork Construction
I think that all things considered, the above code does a great job of getting course information from the HTML. The Course list is modular enough to stand on its own and be used for other means -- making the network is a secondary analysis step.
# For meaningful coloring
def unique_depts(course_list):
return list(set([x.course_code for x in course_list]))
def col_to_hex(x):
return '#{0:02x}{1:02x}{2:02x}'.format(max(0, min(int(x[0]*175.0 + 80.0), 255)), max(0, min(int(x[1]*175.0 + 80.0), 255)), max(0, min(int(x[2]*175.0 + 80.0), 255)))# Dumb wrapper block made for show_network.
# Warning: This whole cell of code is inefficient, hacky, and messy.
# A lot of bugfixing and database correction happened here. I'm going to leave it
# as-is for now, since this visualization is tangential to the project.
def add_node_group(data, DG, dept_list, dept_color_map, course_list, courses_pos):
xp = 0
yp = 0
# Add an edge going to every node mentioned, no matter where it's mentioned.
# Ignore the operators, too: [(), and, or]
for x in data:
if len(x) == 7 and x not in DG.nodes() and x[:3] in dept_list:
if courses_pos != None:
xp = courses_pos[x]['x']
yp = courses_pos[x]['y']
DG.add_node(x, shape='box', color=col_to_hex(dept_color_map[x[:3]]), title="No Data!", x=xp, y=yp)
def show_network(course_list, courses_pos = None):
DG = nx.DiGraph()
dept_list = unique_depts(course_list)
# It's been a while since I touched Python. I forgot how fun one-liners like these are.
dept_color_map = dict(zip(dept_list, distinctipy.get_colors(len(dept_list))))
course_map = dict(zip([x.course_id for x in course_list], course_list))
for c in course_list:
if len(c.prerequisites) + len(c.corequisites) + len(c.recommended) + len(c.complements_courses) > 0:
xp = 0
yp = 0
if courses_pos != None:
xp = courses_pos[c.course_id]['x']
yp = courses_pos[c.course_id]['y']
DG.add_node(c.course_id, shape='box', dept=c.course_code, color=col_to_hex(dept_color_map[c.course_code]), title="No Data!", x=xp, y=yp)
add_node_group(c.prerequisites, DG, dept_list, dept_color_map, course_list, courses_pos)
add_node_group(c.corequisites, DG, dept_list, dept_color_map, course_list, courses_pos)
add_node_group(c.recommended, DG, dept_list, dept_color_map, course_list, courses_pos)
add_node_group(c.complements_courses, DG, dept_list, dept_color_map, course_list, courses_pos)
for x in c.prerequisites:
if len(x) == 7:
DG.add_edge(c.course_id, x, title='Prerequisite')
for x in c.corequisites:
if len(x) == 7:
DG.add_edge(c.course_id, x, title='Corequisite')
for x in c.recommended:
if len(x) == 7:
DG.add_edge(c.course_id, x, title='Recommended Knowledge')
for x in c.complements_courses:
if len(x) == 7:
DG.add_edge(c.course_id, x, title='Complements')
# Hey, guess what: AEE3150 references courses that DON'T EXIST. I think they messed up when typing the code?
# this is stupid the data is stupid this whole entire function is stupid
# duuuuuuuuuuuuuuhhhhhhhhhhhhh
DG.nodes['MAE3083']['x'] = DG.nodes['AEE3150']['x']
DG.nodes['MAE3083']['y'] = DG.nodes['AEE3150']['y']
DG.nodes['MAE3161']['x'] = DG.nodes['AEE3150']['x']
DG.nodes['MAE3161']['y'] = DG.nodes['AEE3150']['y']
# You know, maybe there isn't a real database. Maybe it's just a bunch of unlinked plaintext in an excel file.
# That would explain all the bad entries, at least.
for x in DG.nodes():
if x in course_map:
DG.nodes[x]['title'] = course_map[x].course_name
# display with pyvis for interactibility
g=Network(height=900, width=1400, notebook=True, directed=True)
g.force_atlas_2based()
g.set_edge_smooth('continuous')
# Hrm. I'd like physics to be on by default, but then visjs tries to stabilize the network,
# even though I turn that off here. Fix in HTML?
g.toggle_physics(False)
g.toggle_stabilization(courses_pos==None)
g.from_nx(DG) # I do love how friendly Python can be (when you're not installing modules, that is)
g.show_buttons('physics')
g.show("grph.html")
# Wow, am using these comments as a blogging platform?
# Actually, I think I'm just procrastinating writing the PDAG logic.
# beebeebooboo
# okay, fine
return DGprint('parsing...')
CL = parse_html('webpages/descpages/') # This can take a while when run for the first time
print('making network...')
# I exported all the positions so we don't have to wait for the network to stabilize.
# I got them with ForceAtlas2 and some manual dragging of components out of local minima
saved_pos = json.loads(open('full_network_positions.json', 'r').read())
DG = show_network(CL, saved_pos)
print('done.')First Impressions
Let's see what we're working with: 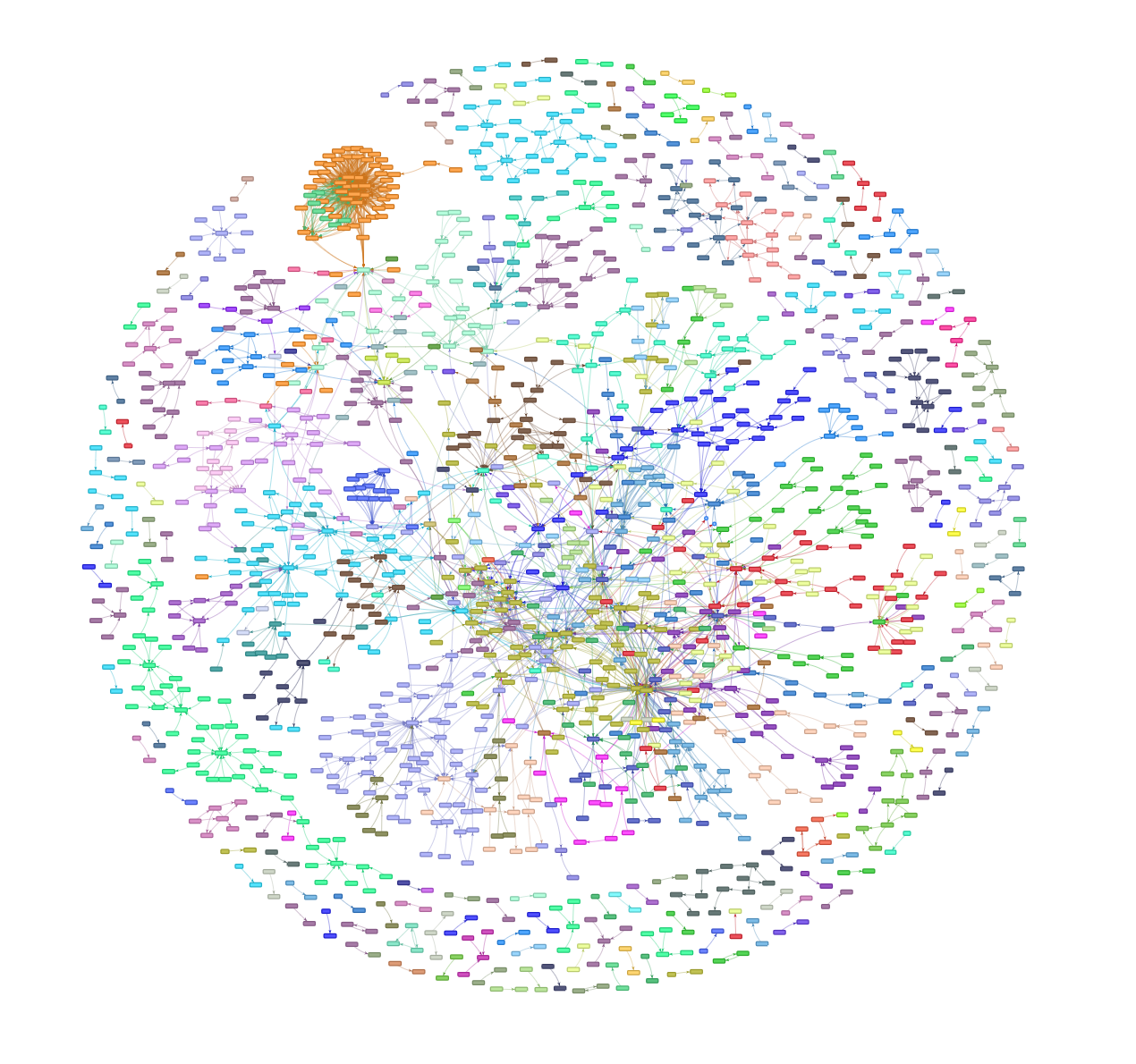 Neat!
Neat!
Nodes are courses, edges are relationships between them. Colors are based on departament codes. Obviously it's a DAG even though the directions aren't visible here. We've got a lot of isolated vertices, plenty of small components, and one massive one. The dense cluster on the left is an interesting anomaly. We'll explore that later. For now, we need to find a way to handle the logical operators in these prerequisite lists -- stuff like (MTH0000 and MTH0001) or MTH0002.
PDAGs (Propositional Directed Acyclic Graphs)
Right now, our edge data looks like this:
[ '(', 'MTH0000', 'and', 'MTH0001', ')', 'or', 'MTH0002']In the image from the previous section, all the logical operators and parenthesis are completely ignored. We're losing data.
Enter PDAGs: a means of representing logical expressions in graphs. The concept is fairly simple: A node for every operand and an edge for every operator. If we generate these graphs for every course, we can compose them with the course network (while allowing them to retain some labels pertaining to their type and function).
This isn't too hard to do, especially since our PDAGs are already simplified past negation normal form -- their operators are limited to just conjunction and disjunction. Also, since both and and or are associative and commutative, we'll use just one or node for cases like MTH1001 or MTH1010 or MTH1702.
Prepping the Data
The dataset has some ambiguous entries that don't use parenthesis despite having multiple operators. Let's introduce an order of operations:
# Issue: not every entry containing multiple operators uses parenthesis (:|).
# Here are all the tricky cases:
# ['BIO5210', 'and', 'BME5300', 'or', 'CHE5300']
# ['(', 'CSE1400', 'or', 'MTH1000', 'or', 'MTH1001', 'or', 'MTH1002', 'or', 'MTH1010', 'or', 'MTH1020',
# 'or', 'MTH1603', 'or', 'MTH1701', 'or', 'MTH1702', 'or', 'MTH2001', 'or', 'MTH2010', 'or', 'MTH2051',
# 'or', 'MTH2201', 'or', 'MTH3200', ')', 'or', '(', 'MTH1011', 'and', 'MTH1012', ')', 'and', 'PSY1411']
# ['BIO4101', 'and', 'BIO4110', 'or', 'BIO4111']
# ['CSE1502', 'or', 'CSE1503', 'or', 'CSE2050', 'and', 'MTH2201']
# ['BME3030', 'and', 'BME5300', 'or', 'CHE5300']
# ['BIO2110', 'or', 'BIO2111', 'and', 'BIO2301']
# ['BIO2110', 'or', 'BIO2111', 'and', 'BIO4010', 'or', 'BIO4011']
# ['CHE1091', 'and', 'CHE3260', 'or', 'CHM2002']
# ['BME3260', 'or', 'CHE3260', 'and', 'AEE3083', 'or', 'BME3081', 'and', 'MEE2024', 'or', 'CHE4568']
# ['MTH1002', 'or', 'MTH1020', 'and', 'BME3260', 'or', 'CHE3260', 'or', 'CSE2410', 'or', 'ECE3551']
# ['CSE1001', 'and', 'MTH2201', 'or', 'MTH3200']
# ['CSE1001', 'and', 'MTH2201', 'or', 'MTH3200']
# In all of these, it seems like OR precedes AND (in order of ops.)
# We'll assume that's the case across the board.
# First, we'll concatenate everything that is already grouped, so we can treat it as one element.
def concat_paren_groups(list_in):
ls = []
# Just gonna roll with a simple FSM here.
in_parens = False
paren_cat = []
for x in list_in:
if in_parens:
if x == ')':
in_parens = False
# This is how I handle nested parenthesis. Since this function is already aggregating
# everything in the parens, it might as well also send it back to the correction function.
# It's sort of superfluous since nested parens never happen in the data, though...
corrected = '~'.join(correct_parens_recur(paren_cat))
ls.append('(~' + corrected + '~)') # the tilde is a temporary substitute for a space
paren_cat = []
else:
paren_cat.append(x)
elif x == '(':
in_parens = True
else:
ls.append(x)
return ls
# Next, we need to identify where to add parenthesis.
# Yes, it does call itself, but it does it through concat_paren_groups()
def correct_parens_recur(list_in):
# loop backwards
ls = list_in.copy()
# deal with any existing grouping
if any('(' in x for x in ls):
ls = concat_paren_groups(ls)
state='nul'
state_ever_changed = False
last_change = len(ls)-1
for i in range(len(ls)-1, 0, -1):
# if we find a different operator at our current position
if ls[i] != state and (ls[i]=='and' or ls[i]=='or'):
# ... and it's not the first real state we find
if not state == 'nul':
# insert parenthesis around any OR set that is on the same hierarchical level as an AND
if state == 'or':
ls.insert(last_change+2, ')')
ls.insert(i+1, '(')
last_change = i
state_ever_changed = True
state = ls[i]
# throw the parens in if we reached the end of the list and the conditions are right
if state_ever_changed and state == 'or':
ls.insert(last_change+2, ')')
ls.insert(0, '(')
return ls
# anything not a course, operator, or parenthesis returns false
def valid_course_data(x):
return bool(re.search('\(|\)|or|and|[A-Z]{3}[0-9]{4}', x))
# changes "( MTH1000 or )" to "( MTH1000 )"
# takes a list of strings as input though, not a string
def remove_degenerate_operators(list_in):
list_out = []
was_op = False
for x in list_in:
list_out.append(x)
if was_op and x == ')':
del list_out[-2]
was_op = ('or' in x.lower() or 'and' in x.lower())
return list_out
# changes "( ( ( MTH1000 ) ) ) and MTH1001" to "MTH1000 and MTH1001"
# takes a list of strings as input though, not a string
def remove_degenerate_parens(list_in):
list_out = []
list_tmp = list_in.copy()
# loop so we get all the nested parenthesis
# there is a way to get them in one pass (keep track of depth), but I don't feel like writing it
has_degen = True
while has_degen:
cells_since_Lparen = 0
to_add = []
list_out = []
has_degen = False
for x in list_tmp:
to_add.append(x)
if x == '(':
cells_since_Lparen = 0
elif x == ')':
# If we see a ')' 2 spaces after a '(', then the parenthesis only enclose
# one value, so we can delete them.
if cells_since_Lparen == 2:
del to_add[-3] # crazy how this works in Python
del to_add[-1]
has_degen = True
list_out.extend(to_add)
to_add = []
cells_since_Lparen += 1
list_out.extend(to_add)
list_tmp = list_out.copy()
return list_out
# wrapper function for correct_parens_recur
# also removes any non-course data and fixes some formatting issues
def fix_formatting(list_in):
# remove degenerates
ls = remove_degenerate_operators(list_in)
ls = remove_degenerate_parens(ls)
ls = correct_parens_recur(ls)
# remove the whitespace placeholders ('~'), and any zero-width space characters ('\u200b')
ls = re.sub('\u200b', '', re.sub('~', ' ', ' '.join(ls))).split(' ')
# remove any 'background knowledge' or similar non-parsable entries
ls = list(filter(valid_course_data, ls))
# remove any trailing operators as a result of the previous filter
if len(ls) > 0 and (ls[0] == 'and' or ls[0] == 'or'): del ls[0]
if len(ls) > 0 and (ls[-1] == 'and' or ls[-1] == 'or'): del ls[-1]
# get rid of operators and do another paren pass
ls = remove_degenerate_operators(ls)
ls = remove_degenerate_parens(ls)
return lsAlgorithm Time
The purpose of this function is to create a small NetworkX structure that can be overlayed on our complete network, or a subset of it. It needs to generate PDAGs from a logical expression in infix notation. This expression may contain nested parenthesis, but its operators are limited to and and or. This task is somewhat analogous to that of an operator precedence parser.
# We'll use a recursive function
def parse_conns_recur(ls, root_node_name, edge_label='???'):
DG = nx.DiGraph()
node_type = 'NULL'
if len(ls) == 0:
DG.add_node(root_node_name)
return DG
if len(ls) == 1:
# this case is only triggered if the root node has no logic
DG.add_node(ls[0])
DG.add_edge(root_node_name, ls[0], title=edge_label)
else:
in_parens = 0
paren_cat = []
to_add = []
to_add_parens = []
for x in ls:
if x == '(':
in_parens += 1
if in_parens > 0:
paren_cat.append(x)
else:
if len(x) == 7:
to_add.append(x)
else:
node_type = x
if x == ')':
in_parens -= 1
if in_parens == 0:
to_add_parens.append(paren_cat[1:-1])
paren_cat = []
if len(to_add) > 0 or len(to_add_parens) > 0:
DG.add_node(root_node_name)
this_node_name = ' '.join(ls)
DG.add_node(this_node_name, label=node_type.upper())
DG.add_edge(root_node_name, this_node_name, title=edge_label)
if len(to_add) > 0:
for x in to_add:
DG.add_node(x)
DG.add_edge(this_node_name, x, title=edge_label)
if len(to_add_parens) > 0:
for x in to_add_parens:
SG = parse_conns_recur(x, this_node_name, edge_label)
DG = nx.compose(DG, SG)
return DG
# Another wrapper to correct the parenthesis in our input list
def get_PDAG(ls, root_node_name, edge_label):
filt_list = fix_formatting(ls)
DG = parse_conns_recur(filt_list, root_node_name, edge_label)
return DGDisplaying the Network (again)
We'll display it like how we did before, but a little neater this time.
# SG for... uh... supergraph?
SG = nx.DiGraph()
# this can take a few seconds
for x in CL:
DG2 = get_PDAG(x.prerequisites, x.course_id, 'prerequisite')
SG = nx.compose(SG, DG2)
# complements_courses has no logical operators, it's just a list of courses, so we won't pass it to parse_connsdept_list = unique_depts(CL)
# It's been a while since I touched Python. I forgot how fun one-liners like these are.
dept_color_map = dict(zip(dept_list, distinctipy.get_colors(len(dept_list), pastel_factor=0.85)))
def disp_PDAG(pdag_graph, remove_isolates=True):
gn=Network(height=900, width=1400, notebook=False, directed=True)
for x in pdag_graph.nodes():
pdag_graph.nodes[x]['shape'] = 'box'
if 'label' in pdag_graph.nodes[x]:
if pdag_graph.nodes[x]['label'] == 'OR':
pdag_graph.nodes[x]['label'] = ' '
pdag_graph.nodes[x]['color'] = '#ffaaaa'
pdag_graph.nodes[x]['shape'] = 'triangleDown'
if pdag_graph.nodes[x]['label'] == 'AND':
pdag_graph.nodes[x]['label'] = ' '
pdag_graph.nodes[x]['color'] = '#aaffaa'
pdag_graph.nodes[x]['shape'] = 'triangle'
else:
code = x[0:3]
if code in dept_color_map:
pdag_graph.nodes[x]['color'] = col_to_hex(dept_color_map[code])
pdag_graph.remove_nodes_from(list(nx.isolates(pdag_graph)))
gn.force_atlas_2based()
gn.set_edge_smooth('continuous')
gn.toggle_physics(False)
gn.from_nx(pdag_graph)
gn.show_buttons('physics')
gn.show("grph_pdag.html")
disp_PDAG(SG)Second Impressions
Note: You can view an interactive version of the following network here. The network (this time of just prerequisites) has roughly the same large-scale structure:
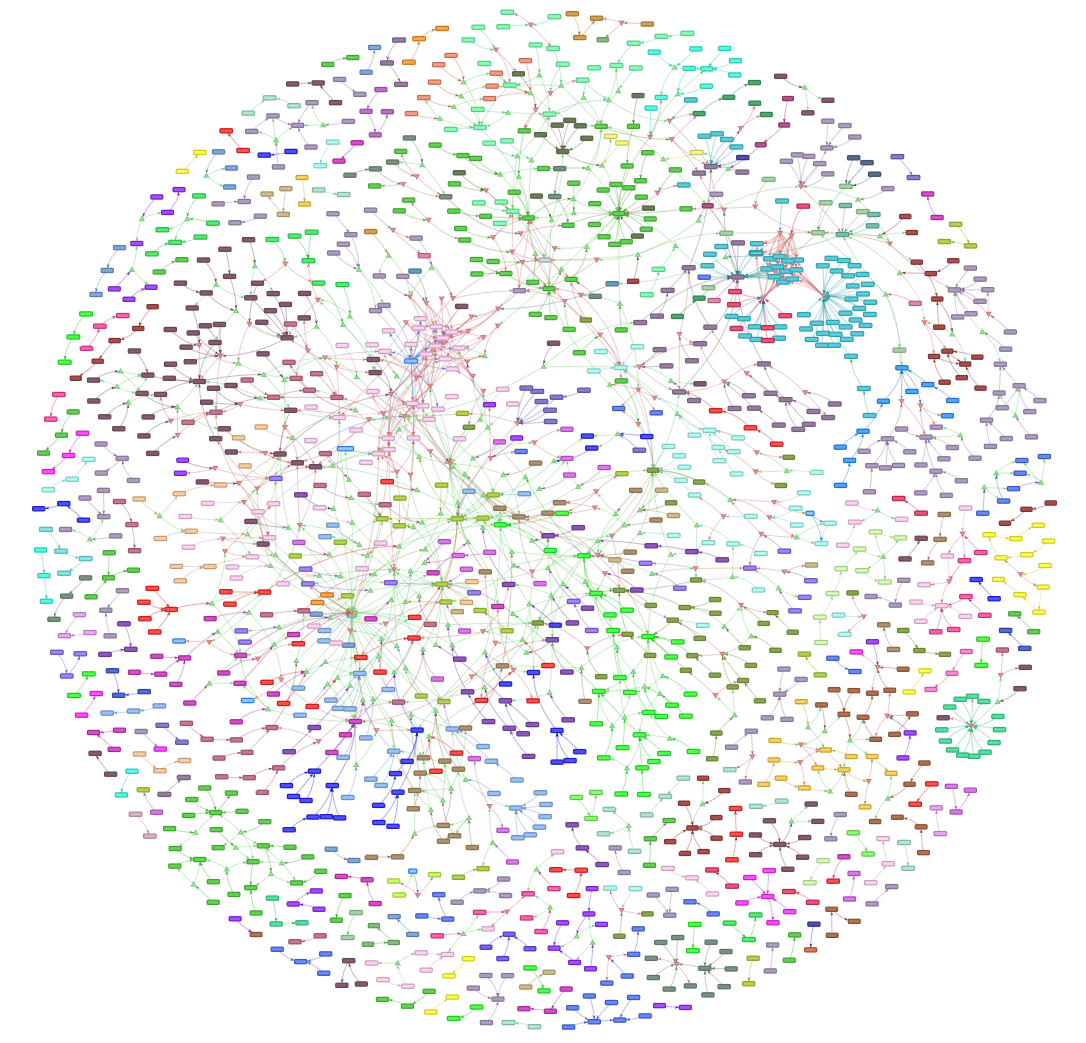
But if we zoom in, we can see there's a lot more going on at small scales.
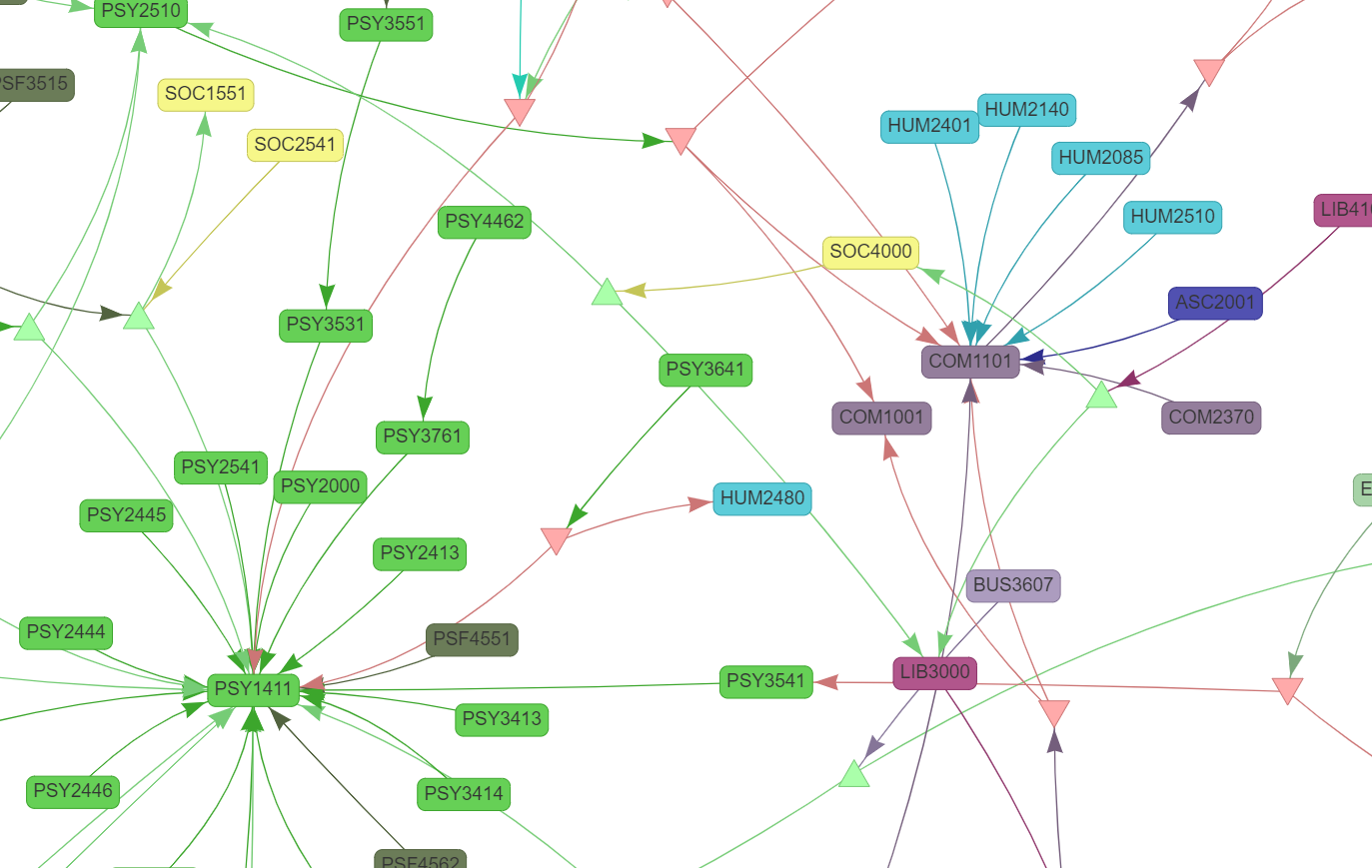
The red, downward-facing triangles represent an OR operator, and the green upward-facing triangles represent an AND operator. With this, the network is now able to fully represent the logic in the course catalog data.
Continued in Part II
Previous Post:Upgrading Florida Tech's CAPP Reports (Part 2) Next Post:
The Chromatic Aberration Rant